Federated Learning for Medical Imaging
Published:
Nearly 153 exabytes of healthcare-related data were generated in 2013; this number will increase by 48% annually to reach 2,314 exabytes in 2020 [1], [2], [3]. While machine learning can benefit from this “big data” to generate state-of-the-art models, most healthcare data is hard to obtain due to legal, privacy, technical, and data-ownership challenges, especially among international institutions where HIPAA and GDPR concerns need to be addressed [3], [4].
Federated learning, introduced by Google in 2017, is a distributed machine learning approach that enables multi-institutional collaboration on deep learning projects without sharing patient data. In 2018, Intel began a collaboration with the Center for Biomedical Image Computing and Analytics (CBICA) at the University of Pennsylvania to show the first proof-of-concept application of federated learning to real-world medical imaging [5] (Figure 1). Our initial study demonstrated that federated learning could train a deep learning model (U-Net, [10]) to 99% of the accuracy of the same model trained with the traditional data-sharing method (Figures 2 and 3). In September, we presented our results at the Medical Image Computing and Computer Assisted Intervention (MICCAI) in Granada, Spain. We recently published our results in the Springer’s Lecture Notes in Computer Science [5].
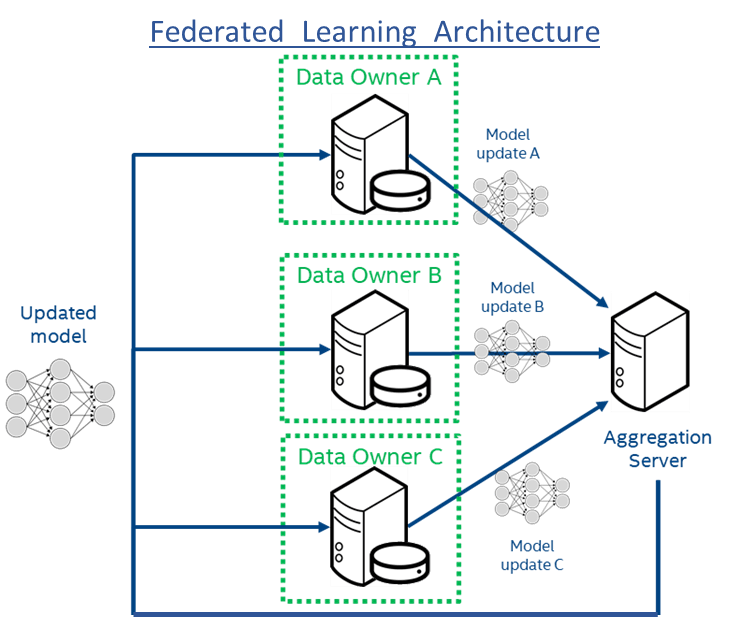
Figure 1: Federated Learning Architecture using Intel hardware. The encrypted model is sent to the individual institutions (Data Owners A-C) which decrypt within a secure enclave in hardware and then train on the local data. Only the model updates are shared with the central model aggregator. This provides protection to both the model and the data. The raw data never leaves the institutions, which not only adds privacy but also prevents large data transfers on the network.
Currently, the University of Pennsylvania and 19 other institutions worldwide are leading the first real-world medical use case of federated learning. Intel will provide support to the project by leveraging the capabilities of our Intel® Xeon® Scalable processors and Intel® Software Guard Extensions (Intel® SGX). We will show how Intel technology can enhance the security of federated learning by protecting both the data and the model being trained. Our hope is that Intel can provide researchers with the technology to create solutions for federated learning that will enable generalizable, state-of-the-art healthcare models while increasing the protection of sensitive patient data. Learn more about our AI initiatives in health & life sciences and follow us to get the latest AI news from Intel.
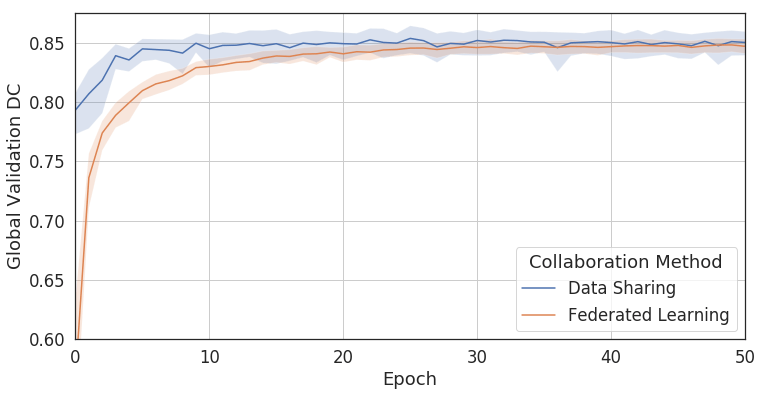
Figure 2: Comparing Federated Learning to data sharing. Training a convolutional neural network (U-Net, [10]) with Federated Learning achieves 99% of the accuracy without sharing patient data [cf. [5]].
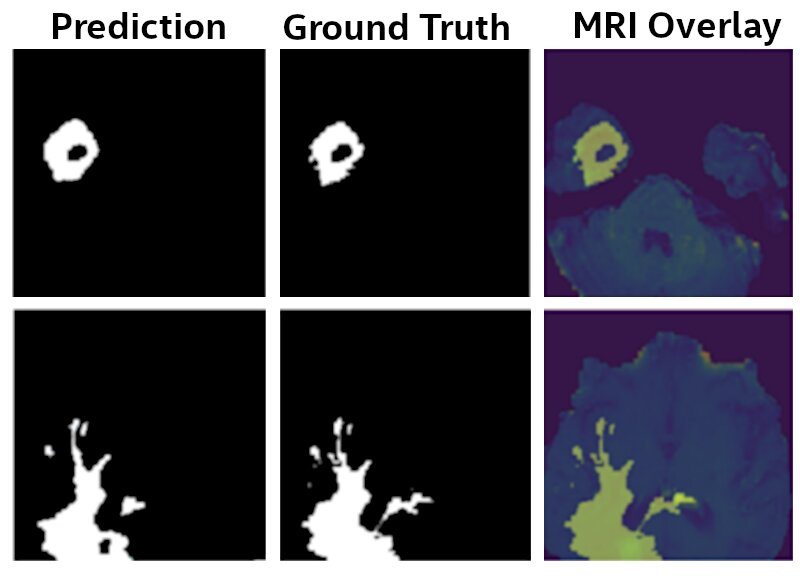
Figure 3: U-Net Model results. The final model identifies Glioma brain tumors from MRI scans with 99% of the accuracy as a model that was trained by sharing the raw MRI data, as provided by the BraTS initiative [6-9].
Intel is partnering with the University of Pennsylvania and 19 other medical research institutions on development of a secure federated learning platform, which will enable collaborators to train a shared machine learning model for healthcare without exchanging confidential patient data.
References
[1] Corbin K. How CIOs Can Prepare for Healthcare “Data Tsunami” [Internet]. CIO. 2014 [cited 8 FEB 2019].
[2] Fenton SH, Low S, Abrams KJ, Butler-Henderson K. Health Information Management: Changing with Time. IMIA Yearbook of Medical Informatics 2017.
[3] Stanford Medicine. 2017 Health Trends Report: Harnessing the Power of Data in Health. Accessed online 8 FEB 2019.
[4] Cho J, Lee K, Shin E, Choy G, Do S. How much data is needed to train to a medical image deep learning system to achieve necessary high accuracy? ICLR 2016.
[5] Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. Lecture Notes in Computer Science book series (Volume 11383). 2019.
[6] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. “Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features”, Nature Scientific Data, 4:170117 (2017) DOI: 10.1038/sdata.2017.117.
[7] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C. “Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-GBM collection”, The Cancer Imaging Archive, 2017. DOI: 10.7937/K9/TCIA.2017.KLXWJJ1Q.
[8] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C. “Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-LGG collection”, The Cancer Imaging Archive, 2017. DOI: 10.7937/K9/TCIA.2017.GJQ7R0EF.
[9] Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Γ, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. “The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)”, IEEE Transactions on Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694
[10] Ronneberger O., Fischer P. , and T. Brox. “U-Net Convolutional Networks for Biomedical Image Segmentation.” arXiv:1505.04597v1 [cs.CV] 18 May 2015.